2.3 GWAS
It’s required to provide genotype, map (if applicable), and phenotype files to run GWAS in iPat. Covariates and kinship matrix are optional. User-provided covariates will serve as fixed effects in the GWAS model. Often time users are not recommended to provide a kinship matrix, since those implemented tools will generate one automatically and user-define kinship is used only in ‘MLM’ and ‘CMLM’ from GAPIT.
Output files from GWAS include:
- Figures: Manhattan plot, Q-Q plot, Heterozygosity distribution, and Phenotype overview
- Tables: GWAS report
2.3.1 GAPIT
## Column 1 ['V1'] of item 2 is missing in item 1. Use fill=TRUE to fill with NA (NULL for list columns), or use.names=FALSE to ignore column names. use.names='check' (default from v1.12.2) emits this message and proceeds as if use.names=FALSE for backwards compatibility. See news item 5 in v1.12.2 for options to control this message.
## Column 1 ['V1'] of item 2 is missing in item 1. Use fill=TRUE to fill with NA (NULL for list columns), or use.names=FALSE to ignore column names. use.names='check' (default from v1.12.2) emits this message and proceeds as if use.names=FALSE for backwards compatibility. See news item 5 in v1.12.2 for options to control this message.| Arguments | Default | Options | Description |
|---|---|---|---|
| Model | GLM | GLM, MLM, CMLM, FarmCPU | Implemented models: general linear model (GLM), mixed linear model (MLM), compressed mixed linear model (CMLM), and FarmCPU. |
| PC | 3 | Ranges between 3 to 10 | Principal components to control spurious signals caused by population stratifications |
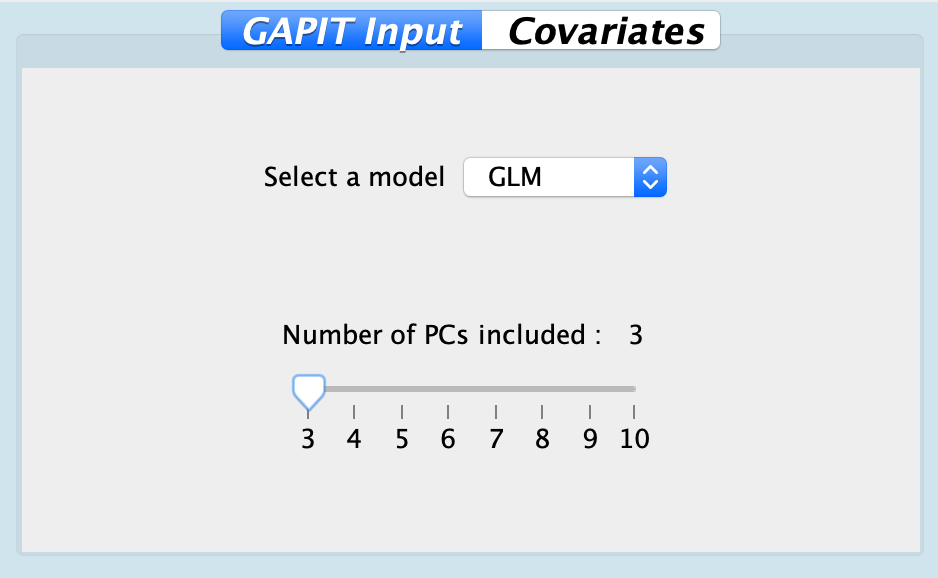
Fig. Configuration for GAPIT
Reference
- Tang,Y. et al. (2016) GAPIT Version 2: An Enhanced Integrated Tool for Genomic Association and Prediction. Plant J., 9.
2.3.2 FarmCPU
## Column 1 ['V1'] of item 2 is missing in item 1. Use fill=TRUE to fill with NA (NULL for list columns), or use.names=FALSE to ignore column names. use.names='check' (default from v1.12.2) emits this message and proceeds as if use.names=FALSE for backwards compatibility. See news item 5 in v1.12.2 for options to control this message.
## Column 1 ['V1'] of item 2 is missing in item 1. Use fill=TRUE to fill with NA (NULL for list columns), or use.names=FALSE to ignore column names. use.names='check' (default from v1.12.2) emits this message and proceeds as if use.names=FALSE for backwards compatibility. See news item 5 in v1.12.2 for options to control this message.| Arguments | Default | Options | Description |
|---|---|---|---|
| Method bin | optimum | static, optimum | |
| maxLoop | 10 | Ranges between 1 to 10 | Number of iterations for FarmCPU to detect QTNs |
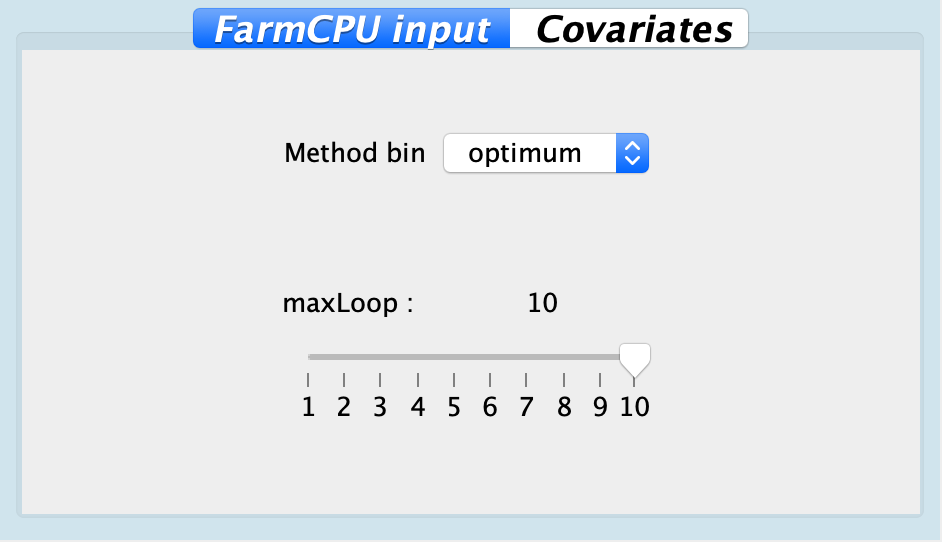
Fig. Configuration for FarmCPU
Reference
- Liu,X. et al. (2016) Iterative Usage of Fixed and Random Effect Models for Powerful and Efficient Genome-Wide Association Studies. PLoS Genet., 12 , e1005767
2.3.3 PLINK
## Column 1 ['V1'] of item 2 is missing in item 1. Use fill=TRUE to fill with NA (NULL for list columns), or use.names=FALSE to ignore column names. use.names='check' (default from v1.12.2) emits this message and proceeds as if use.names=FALSE for backwards compatibility. See news item 5 in v1.12.2 for options to control this message.
## Column 1 ['V1'] of item 2 is missing in item 1. Use fill=TRUE to fill with NA (NULL for list columns), or use.names=FALSE to ignore column names. use.names='check' (default from v1.12.2) emits this message and proceeds as if use.names=FALSE for backwards compatibility. See news item 5 in v1.12.2 for options to control this message.| Arguments | Default | Options | Description |
|---|---|---|---|
| C.I. | 0.95 | 0.95, 0.975, 0.995 | Confident interval for testing marker effects |
| Method | GLM | GLM, Logistic Regression | Implemented models |
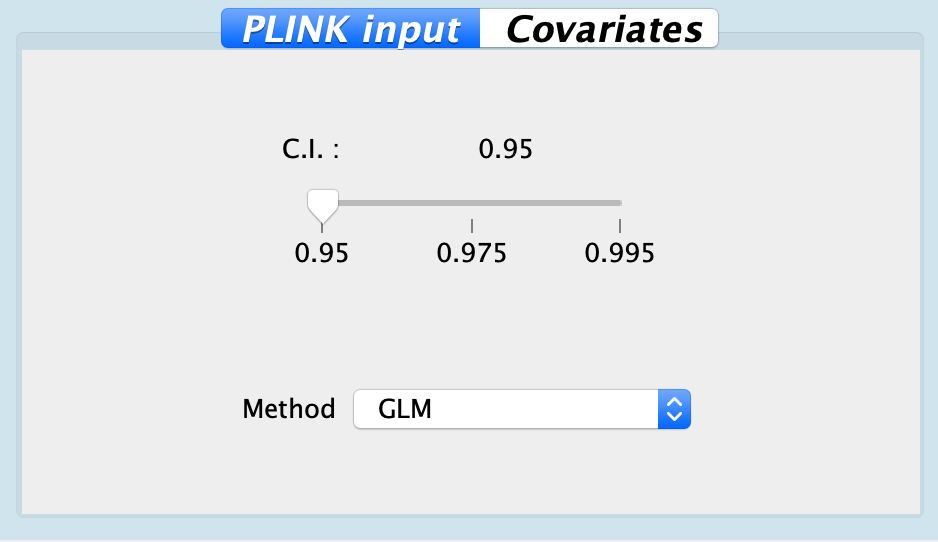
Fig. Configuration for PLINK
Reference
- Purcell,S. et al. (2007) PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am J Hum Genet, 81, 559–575.