2.2 Global settings
2.2.1 Working directory
Users can specify the working directory and module names in this panel. Module names will be presented in the output, which can serve as a label for users to distinguish different analyses.
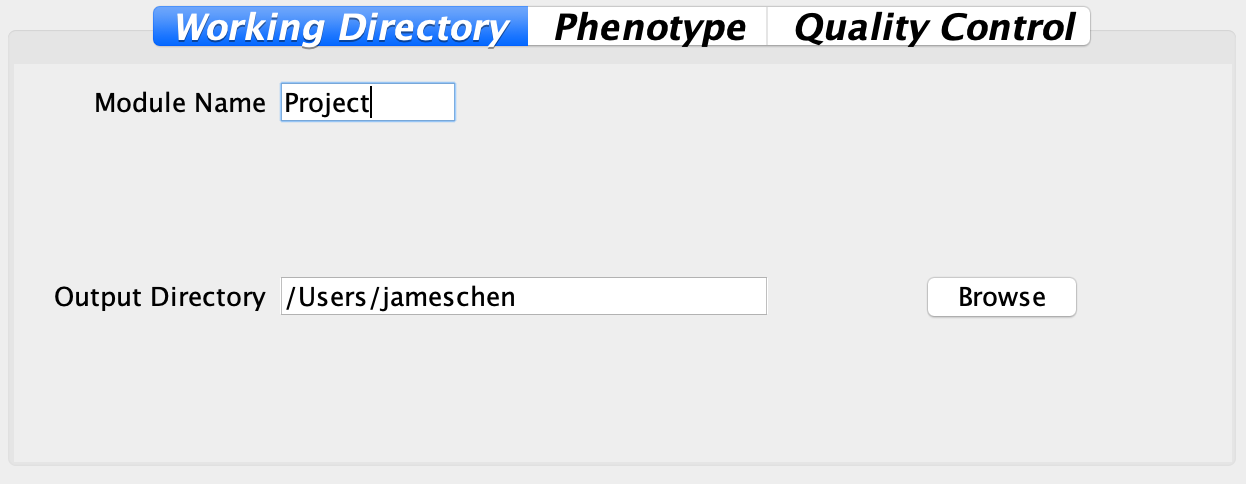
Fig. Panel for users to define working directory
2.2.2 Phenotype
Sometimes users may have several traits in one phenotype file, but not all of them are traits of interest. To deal with this concern, in this panel, traits can be selected and excluded for the following analysis. Click ‘–>’ button to exclude traits that are not the interest of this analysis, or click ‘<–’ button to rescue back traits that are mistakenly excluded.
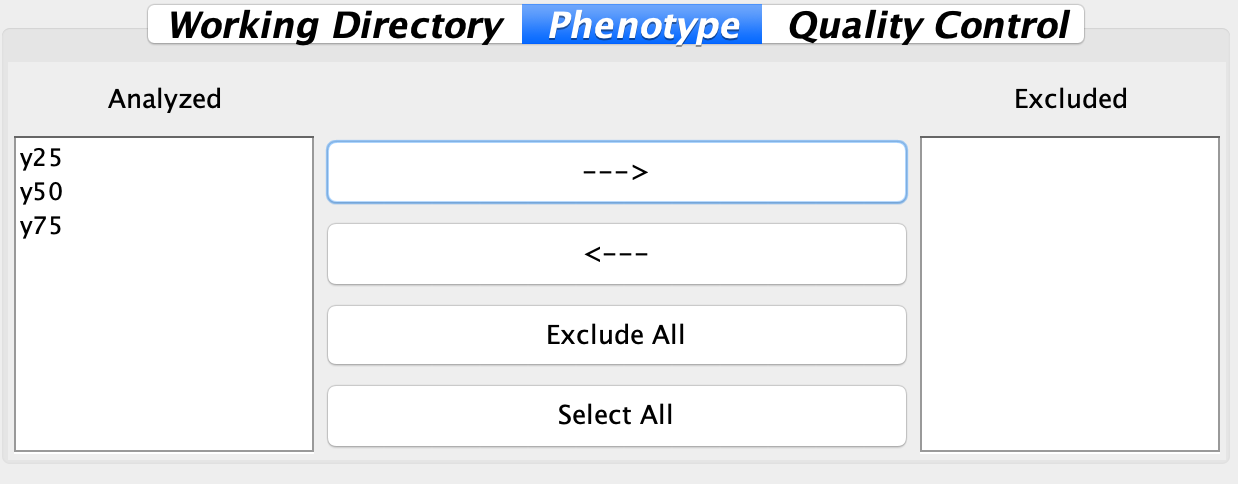
Fig. Panel for users to select traits of interest
2.2.3 Quality Control
Two ways of quality controls (QC) are available in this panel, both ways will be applied to genetic markers. ‘By missing rate’ will filter out any marker that has missing rate (i.e., 1 - calling rate) higher than the assigned value. ‘By MAF’ will filter out markers with minor allele frequency lower than the assigned value.

Fig. Sliders for performing quality controls