Customize Vegetation Indices
Objectives
Derive a index that can better associate with the canopy area of alfalfa than NDVI does. The input image used in this tutorial is an orthoimage with 6 spectral channels. We will demonstrate how to utilize the output files to perform a simple feature selection.
Inspect output files
[1]:
# imports
import os
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
from scipy.stats import pearsonr
[2]:
# inspect the output files
os.listdir()
[2]:
['GRID_bin.png',
'GRID_seg_border.png',
'GRID.shx',
'.DS_Store',
'GRID.shp',
'GRID_data.csv',
'GRID.dbf',
'GRID_image.npy',
'GRID_seg_ID.png',
'.ipynb_checkpoints',
'GRID_kmeans.png',
'GRID_raw.png']
[3]:
# inspect the input image
img = mpimg.imread('GRID_raw.png')
plt.figure(figsize=(8, 8))
plt.imshow(img)
[3]:
<matplotlib.image.AxesImage at 0x7f94b1002e90>
[4]:
# inspect the tabular output
dt = pd.read_csv("GRID_data.csv"); dt
[4]:
| var | row | col | area_all | area_veg | NDVI | NDVI_std | GNDVI | GNDVI_std | CNDVI | ... | ch_2 | ch_2_std | ch_3 | ch_3_std | ch_4 | ch_4_std | ch_5 | ch_5_std | ch_6 | ch_6_std | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | unnamed_0 | 1 | 1 | 14910 | 6205 | 0.680722 | 0.101848 | 0.481553 | 0.086146 | 0.955003 | ... | 59.423529 | 15.689919 | 26.239484 | 8.893769 | 167.117808 | 21.852656 | 69.889927 | 10.829972 | 30.540088 | 24.633806 |
| 1 | unnamed_23 | 1 | 2 | 10251 | 6074 | 0.676454 | 0.094532 | 0.475063 | 0.077119 | 0.942506 | ... | 59.718966 | 14.534593 | 26.431841 | 8.389850 | 165.603721 | 20.710229 | 69.595324 | 9.956617 | 26.050244 | 23.631348 |
| 2 | unnamed_46 | 1 | 3 | 12996 | 8037 | 0.679914 | 0.089733 | 0.474278 | 0.074406 | 0.944246 | ... | 60.750404 | 14.811752 | 26.750280 | 8.541490 | 167.954585 | 21.471309 | 69.989051 | 10.539928 | 31.216141 | 24.766432 |
| 3 | unnamed_69 | 1 | 4 | 11100 | 5435 | 0.643977 | 0.114235 | 0.442635 | 0.088716 | 0.879109 | ... | 65.185097 | 15.769556 | 28.936155 | 9.405345 | 166.668813 | 21.403908 | 68.871757 | 10.314017 | 20.444095 | 21.586253 |
| 4 | unnamed_92 | 1 | 5 | 12506 | 5645 | 0.659209 | 0.118899 | 0.460301 | 0.092747 | 0.913120 | ... | 62.443933 | 16.140226 | 27.421612 | 9.592832 | 166.453853 | 19.131734 | 69.195748 | 9.295681 | 17.890222 | 21.159997 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 271 | unnamed_183 | 23 | 8 | 13851 | 6455 | 0.743537 | 0.114225 | 0.598427 | 0.093205 | 1.154553 | ... | 44.106274 | 12.926948 | 20.407901 | 8.120920 | 173.637180 | 21.825689 | 72.545778 | 11.594452 | 38.396902 | 19.955167 |
| 272 | unnamed_206 | 23 | 9 | 12877 | 6293 | 0.735538 | 0.113924 | 0.594051 | 0.090958 | 1.140933 | ... | 44.571746 | 13.482742 | 21.143334 | 8.533524 | 172.578738 | 20.879160 | 72.617670 | 11.297813 | 34.843954 | 19.919131 |
| 273 | unnamed_229 | 23 | 10 | 12880 | 7028 | 0.751853 | 0.099948 | 0.605144 | 0.079713 | 1.168800 | ... | 43.526323 | 11.750768 | 20.231929 | 7.710727 | 175.287137 | 22.499670 | 74.050370 | 12.060664 | 40.409932 | 21.961529 |
| 274 | unnamed_252 | 23 | 11 | 13122 | 6433 | 0.734485 | 0.123072 | 0.590896 | 0.098009 | 1.137487 | ... | 45.554018 | 14.221262 | 21.560547 | 9.116043 | 174.559304 | 22.556524 | 73.487331 | 12.153618 | 44.907508 | 16.025357 |
| 275 | unnamed_275 | 23 | 12 | 15252 | 7576 | 0.740249 | 0.106438 | 0.593371 | 0.087980 | 1.144174 | ... | 45.191526 | 12.338402 | 21.199842 | 8.073755 | 175.554250 | 23.091342 | 74.060058 | 12.693901 | 43.955649 | 18.627713 |
276 rows × 29 columns
[5]:
# subset columns of canopy area, NDVI, and spectral signals
dt_idx = dt[["area_veg", "NDVI"] + ["ch_%d" % i for i in range(1, 7)]].dropna(); dt_idx
[5]:
| area_veg | NDVI | ch_1 | ch_2 | ch_3 | ch_4 | ch_5 | ch_6 | |
|---|---|---|---|---|---|---|---|---|
| 0 | 6205 | 0.680722 | 32.572442 | 59.423529 | 26.239484 | 167.117808 | 69.889927 | 30.540088 |
| 1 | 6074 | 0.676454 | 32.683240 | 59.718966 | 26.431841 | 165.603721 | 69.595324 | 26.050244 |
| 2 | 8037 | 0.679914 | 32.748165 | 60.750404 | 26.750280 | 167.954585 | 69.989051 | 31.216141 |
| 3 | 5435 | 0.643977 | 36.915731 | 65.185097 | 28.936155 | 166.668813 | 68.871757 | 20.444095 |
| 4 | 5645 | 0.659209 | 35.175554 | 62.443933 | 27.421612 | 166.453853 | 69.195748 | 17.890222 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 271 | 6455 | 0.743537 | 26.120372 | 44.106274 | 20.407901 | 173.637180 | 72.545778 | 38.396902 |
| 272 | 6293 | 0.735538 | 27.076593 | 44.571746 | 21.143334 | 172.578738 | 72.617670 | 34.843954 |
| 273 | 7028 | 0.751853 | 25.338788 | 43.526323 | 20.231929 | 175.287137 | 74.050370 | 40.409932 |
| 274 | 6433 | 0.734485 | 27.561324 | 45.554018 | 21.560547 | 174.559304 | 73.487331 | 44.907508 |
| 275 | 7576 | 0.740249 | 26.680042 | 45.191526 | 21.199842 | 175.554250 | 74.060058 | 43.955649 |
270 rows × 8 columns
Design index formulas
[6]:
# make a dictionary of formulas
# where x0 and x1 stand for channel signals from two different (same) columns
ls_formula = {"(x0 - x1) / (x0 + x1)": lambda x0, x1: (x0 - x1) / (x0 + x1),
"x0 / x1": lambda x0, x1: x0 / x1,
"(x0 ^ 2) + (x1 ^ 2)": lambda x0, x1: (x0 ** 2) + (x1 ** 2)}
Compute correlations
We can compute Pearson’s correlation coefficients between the canopy areas (i.e., the column of “area_veg”) and derived indices
[15]:
# instantiate pandas dataframe
dt_results = pd.DataFrame(columns=["x0", "x1", "formula", "r2"])
# define the loop
cols_ignored = 2 # we won't loop over the first 2 columns, which are canopy and NDVI
p = 6 # number of channels we'd like to loop over
# pairwise iterate through 6 spectral bands
for i in range(p):
for j in range(p):
# get band signals and store them in x0 and x1
x0 = dt_idx.iloc[:, cols_ignored + i]
x1 = dt_idx.iloc[:, cols_ignored + j]
# compute 3 indices from x0 and x1
ls_idx = [list(ls_formula.values())[i](x0, x1) for i in range(3)]
# store results into a temporary dataframe
dt_temp = pd.DataFrame({
"x0": i,
"x1": j,
"formula": list(ls_formula.keys()),
"r": [pearsonr(dt_idx["area_veg"], ls_idx[i])[0]
for i in range(3)],
"r2": [pearsonr(dt_idx["area_veg"], ls_idx[i])[0] ** 2
for i in range(3)]
})
# append our result dataframe
dt_results = dt_results.append(dt_temp)
[8]:
# handle results from zero division error
dt_results.loc[dt_results.r2.isna(), ["r2", "r"]] = 0
dt_results
[8]:
| x0 | x1 | formula | r2 | r | |
|---|---|---|---|---|---|
| 0 | 0 | 0 | (x0 - x1) / (x0 + x1) | 0.000000 | 0.000000 |
| 1 | 0 | 0 | x0 / x1 | 0.000000 | 0.000000 |
| 2 | 0 | 0 | (x0 ^ 2) + (x1 ^ 2) | 0.230163 | -0.479753 |
| 0 | 0 | 1 | (x0 - x1) / (x0 + x1) | 0.124206 | -0.352429 |
| 1 | 0 | 1 | x0 / x1 | 0.134024 | -0.366092 |
| ... | ... | ... | ... | ... | ... |
| 1 | 5 | 4 | x0 / x1 | 0.010826 | 0.104048 |
| 2 | 5 | 4 | (x0 ^ 2) + (x1 ^ 2) | 0.050595 | 0.224933 |
| 0 | 5 | 5 | (x0 - x1) / (x0 + x1) | 0.000000 | 0.000000 |
| 1 | 5 | 5 | x0 / x1 | 0.000000 | 0.000000 |
| 2 | 5 | 5 | (x0 ^ 2) + (x1 ^ 2) | 0.003587 | 0.059888 |
108 rows × 5 columns
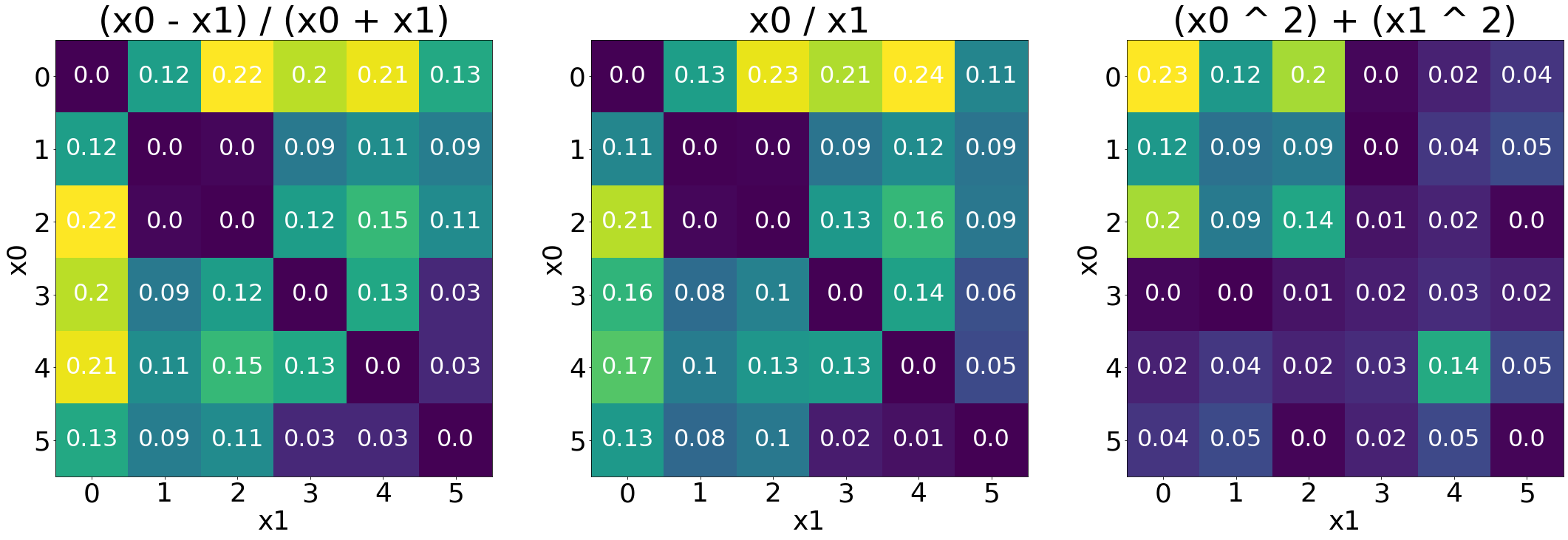
Heatmaps of correlations
This visualization can show how derived indices associate with canopy areas
[9]:
# define font size
size_title = 48
size_label = 32
size_tick = 36
# generate 3 heatmaps for each formula
fig, axes = plt.subplots(1, 3, figsize=(30, 10), constrained_layout=True)
for i in range(3):
# get r2 values by each formula
formula = list(ls_formula.keys())[i]
ls_r2 = dt_results.loc[dt_results.formula == formula, "r2"]
# reshape the list to a 2D matrix
M = np.array(ls_r2).reshape((p, p))
# plot interface
axes[i].set_title(list(ls_formula.keys())[i], fontsize=size_title)
axes[i].set_xlabel("x1", fontsize=size_tick)
axes[i].set_ylabel("x0", fontsize=size_tick)
for x0 in range(p):
for x1 in range(p):
text = axes[i].text(x0, x1, round(M[x1, x0], 2),
ha="center", va="center",
color="w", fontsize=size_label)
plt.sca(axes[i])
plt.xticks(ticks=np.arange(p), fontsize=size_tick)
plt.yticks(ticks=np.arange(p), fontsize=size_tick)
# show heatmap
axes[i].imshow(M)
Tabular Results of selected indices
Also we can use in a dataframe to inspect which indices perform better than NDVI
[10]:
# compute correlation between NDVI and canopy area
r2_ndvi = pearsonr(dt_idx["area_veg"], dt_idx["NDVI"])[0] ** 2
r2_ndvi
[10]:
0.1975636107759065
[11]:
# extract rows containing better indices
dt_results.loc[dt_results.r2 > r2_ndvi].sort_values(by="r2", ascending=False)
[11]:
| x0 | x1 | formula | r2 | r | |
|---|---|---|---|---|---|
| 1 | 0 | 4 | x0 / x1 | 0.238634 | -0.488502 |
| 1 | 0 | 2 | x0 / x1 | 0.230562 | -0.480169 |
| 2 | 0 | 0 | (x0 ^ 2) + (x1 ^ 2) | 0.230163 | -0.479753 |
| 0 | 0 | 2 | (x0 - x1) / (x0 + x1) | 0.220862 | -0.469959 |
| 0 | 2 | 0 | (x0 - x1) / (x0 + x1) | 0.220862 | 0.469959 |
| 0 | 0 | 4 | (x0 - x1) / (x0 + x1) | 0.214008 | -0.462610 |
| 0 | 4 | 0 | (x0 - x1) / (x0 + x1) | 0.214008 | 0.462610 |
| 1 | 2 | 0 | x0 / x1 | 0.213051 | 0.461574 |
| 1 | 0 | 3 | x0 / x1 | 0.209906 | -0.458155 |
| 2 | 0 | 2 | (x0 ^ 2) + (x1 ^ 2) | 0.199044 | -0.446143 |
| 2 | 2 | 0 | (x0 ^ 2) + (x1 ^ 2) | 0.199044 | -0.446143 |
| 0 | 0 | 3 | (x0 - x1) / (x0 + x1) | 0.198422 | -0.445445 |
| 0 | 3 | 0 | (x0 - x1) / (x0 + x1) | 0.198422 | 0.445445 |
Validate the index
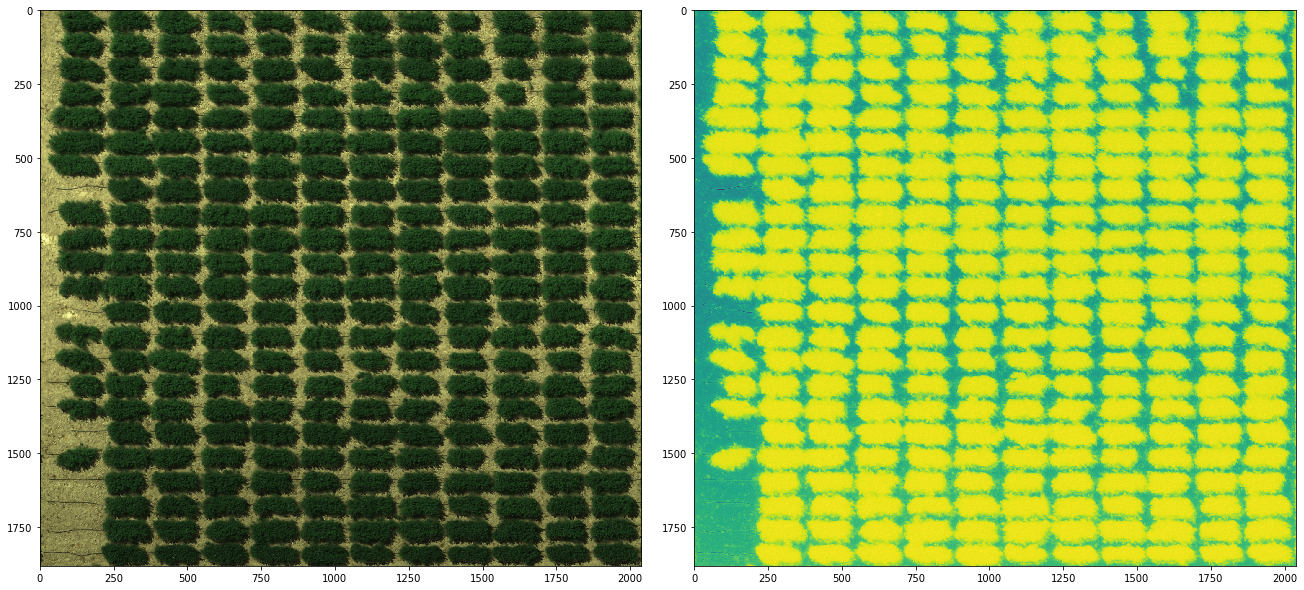
Based on the results shown above, now we know that by dividing the 1st channel by the 5th channel can derive the best canopy-tracking index. we can validate this result by visualize the index as an image.
[13]:
# load image
img = np.load("GRID_image.npy")
img_idx = img[:, :, 0] / (img[:, :, 4] + 1e-9) # adding 1e-9 to avoid zero division error
# as we see the negative correlation,
# we have to treat the largest values as the smallest ones,
# and turn the smallest values to the largest values. (I call it "flip")
img_idx_flip = abs(img_idx - img_idx.max())
# and we can compare the result with the RGB image
fig, axes = plt.subplots(1, 2, figsize=(18, 8), constrained_layout=True)
axes[0].imshow(img[:, :, :3])
axes[1].imshow(img_idx_flip)
[13]:
<matplotlib.image.AxesImage at 0x7f94d05a9c10>